Getting Started¶
Readfish is an adaptive sampling implementation for ONT sequencers. So to begin with you will need an ONT sequencer, currently we support the MinION Mk1B, GridION, and PromethION platforms. As we use live base-calling and alignment the computer that is controlling the sequencer will also need to be capable of base-calling; ideally this would use a GPU.
We recommend for alignment that you pre-prepare a minimap2 index (.mmi) file. This can be done by using the minimap -d flag. An example command -
minimap2 -d <output_index>.mmi <input_reference>.fasta
Where output_index is the name of the index file to be output and input_reference is the name of the FASTA file that you want indexing.
This is then provided as the fn_idx_in value of the TOML file.
Testing your computer¶
To test readfish on your configuration we recommend first running a playback experiment to test unblock speed and then selection.
The following steps should all happen with a configuration (test) flow cell inserted into the target device. A simulated device can also be created within MinKNOW, following these instructions. This assumes that you are runnning MinKNOW locally, using default ports. If this is not the case a developer API token is required on the commands as well, as well as setting the correct port.
If no test flow cell is available, a simulated device can be created within MinKNOW, following the below instructions.
Adding a simulated position for testing
Linux
In the readfish virtual environment we created earlier:
See help
python -m minknow_api.examples.manage_simulated_devices --helpAdd Minion position
python -m minknow_api.examples.manage_simulated_devices --add MS00000Add PromethION position
python -m minknow_api.examples.manage_simulated_devices --prom --add S0Mac
In the readfish virtual environment we created earlier:
See help
python -m minknow_api.examples.manage_simulated_devices --helpAdd Minion position
python -m minknow_api.examples.manage_simulated_devices --add MS00000Add PromethION position
python -m minknow_api.examples.manage_simulated_devices --prom --add S0
As a back up it is possible to restart MinKNOW with a simulated device. This is done as follows:
Stop
minknowOn Linux:
cd /opt/ont/minknow/bin sudo systemctl stop minknow
Start MinKNOW with a simulated device
On Linux
sudo ./mk_manager_svc -c /opt/ont/minknow/conf --simulated-minion-devices=1 &
You may need to add the host 127.0.0.1 in the MinKNOW UI.
Configuring bulk FAST5 file Playback
Download an open access bulk FAST5 file, either R9.4.1 4khz or R10 (5khz). This file is 21Gb so make sure you have sufficient space. A promethION bulkfile is also available but please note this is R10.4 4khz and so will give slightly unexpected results on MinKNOW which assumes 5khz. This file is approx 35Gb in size.
Previously to set up Playback using a pre-recorded bulk FAST5 file, it was necessary to edit the sequencing configuration file that MinKNOW uses. This is currently no longer the case. The “old method” steps are left after this section for reference only or if the direct playback from a bulk file option is removed in future.
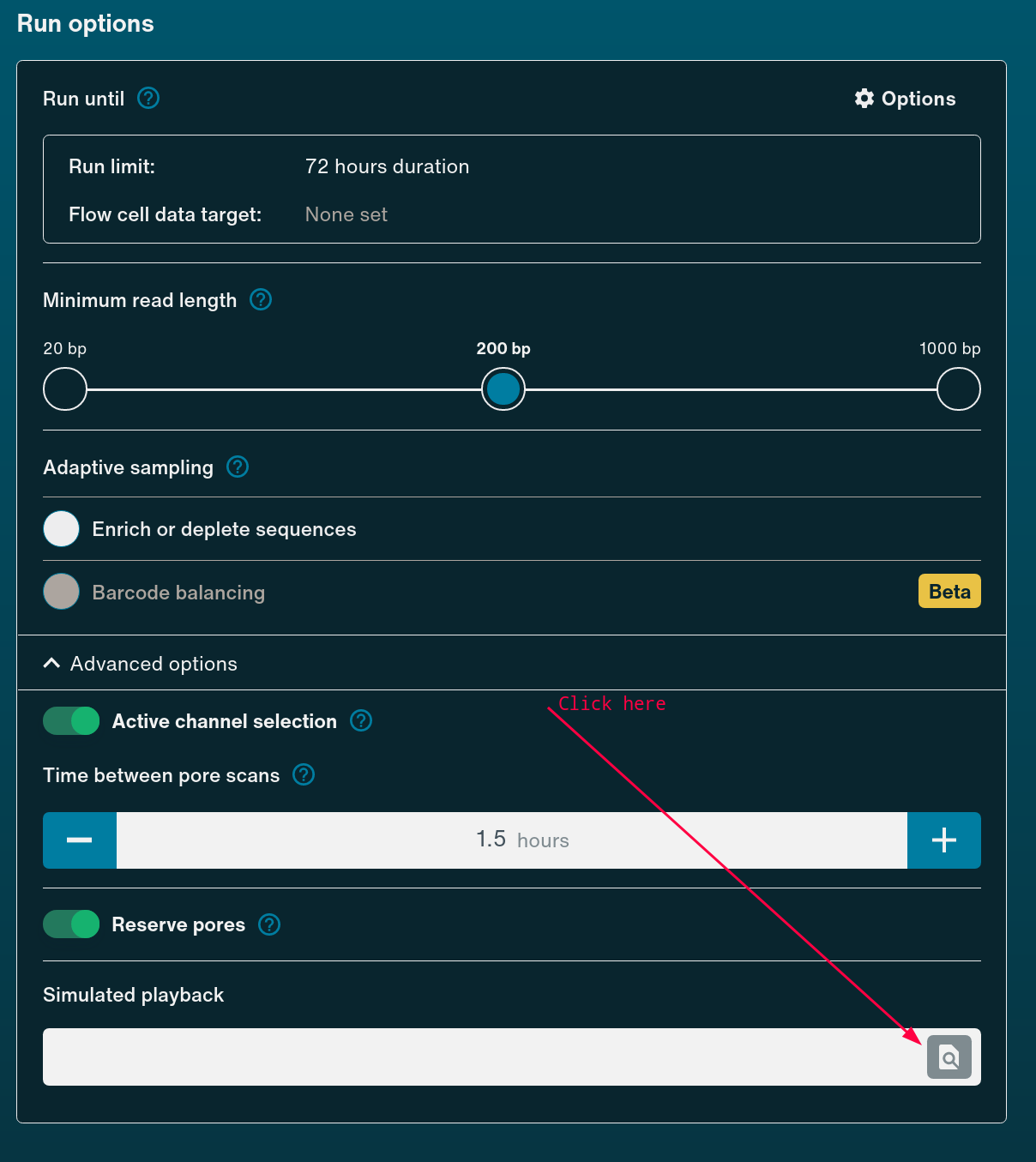
To start sequencing using playback, simply begin setting up the run in the MinKNOW UI as you would usually. Under Run Options you can select Simulated Playback and browse to the downloaded Bulk Fast5 file.
[!NOTE]
Note - The below instructions, whilst they will still work, are no longer required. They are left here for reference only. As of Minknow 5.7, it is possible to select a bulk FAST5 file for playback in the MinKNOW UI.
Old method Configuring bulk FAST5 file Playback
To setup a simulation the sequencing configuration file that MinKNOW uses must be edited.
Steps:
Download an open access bulkfile - either R9.4.1 or R10 (5khz). These files are approximately 21Gb so make sure you have plenty of space. The files are from NA12878 sequencing data using either R9.4.1 or R10.4 pores. Data is not barcoded and the libraries were ligation preps from DNA extracted from cell lines.
A promethION bulkfile is also available but please note this is R10.4, 4khz, and so will give slightly unexpected results on MinKNOW which assumes 5khz.
Copy a sequencing TOML file to the
user_scriptsfolder:On Mac if your MinKNOW output directory is the default:
mkdir -p /Library/MinKNOW/data/user_scripts/simulations cp /Applications/MinKNOW.app/Contents/Resources/conf/package/sequencing/sequencing_MIN106_DNA.toml /Library/MinKNOW/data/user_scripts/simulations/sequencing_MIN106_DNA_sim.toml
On Linux:
sudo mkdir -p /opt/ont/minknow/conf/package/sequencing/simulations cp /opt/ont/minknow/conf/package/sequencing/sequencing_MIN106_DNA.toml /opt/ont/minknow/conf/package/sequencing/simulations/sequencing_MIN106_DNA_sim.toml
Edit the copied file to add the following line under the line that reads “
[custom_settings]”:simulation = "/full/path/to/your_bulk.FAST5"
Change the text between the quotes to point to your downloaded bulk FAST5 file.
Optional, If running Dorado in GPU mode, you can set the parameter
break_reads_after_seconds = 1.0tobreak_reads_after_seconds = 0.4. This results in a smaller read chunk. For R10.4 this is not required but can be tried. For adaptive sampling on PromethION, this should be left at 1 second.In MinKNOW >= 6.0.0 this value defaults to 0.8 which is a reasonable balance.
In the MinKNOW GUI, right click on a sequencing position and select
Reload Scripts. Your version of MinKNOW will now playback the bulkfile rather than live sequencing.Start a sequencing run as you would normally, selecting the corresponding flow cell type to the edited script (here FLO-MIN106) as the flow cell type.
Whichever instructions you followed, the run should start and immediately begin a mux scan. Let it run for around
five minutes after which your read length histogram should look as below:

Testing unblock response
Now we shall test unblocking by running readfish unblock-all which will simply eject
every single read on the flow cell.
To do this run:
readfish unblock-all --device <YOUR_DEVICE_ID> --experiment-name "Testing readfish Unblock All"Leave the run for a further 5 minutes and observe the read length histogram. If unblocks are happening correctly you will see something like the below:
 A closeup of the unblock peak shows reads being unblocked quickly:
A closeup of the unblock peak shows reads being unblocked quickly:

If you are happy with the unblock response, move on to testing base-calling.
Note: The plots here are generated from running readfish unblock-all on an Apple Silicon laptop. The unblock response may be faster on a GPU server.
Testing base-calling and mapping
To test selective sequencing you must have access to a dorado basecall server.
and a readfish TOML configuration file.
First make a local copy of the example TOML file:
curl -O https://raw.githubusercontent.com/LooseLab/readfish/master/docs/_static/example_tomls/human_chr_selection.tomlIf on PromethION, edit the
mapper_settings.mappysection to read:[mapper_settings.mappy-rs]If on MinKNOW core>=5.9.0 and Dorado server version >=7.3.9, edit the
basecallersection to read:[caller_settings.dorado]Modify the
fn_idx_infield in the file to be the full path to a minimap2 index of the human genome.Modify the
targetsfields for each condition to reflect the naming convention used in your index. This is the sequence name only, up to but not including any whitespace. e.g.>chr1 human chromosome 1would becomechr1. If these names do not match, then target matching will fail.
We can now validate this TOML file to see if it will be loaded correctly.
readfish validate human_chr_selection.toml
Errors with the configuration will be written to the terminal along with a text description of the conditions for the experiment as below.
2023-10-05 15:29:18,934 readfish /home/adoni5/mambaforge/envs/readfish_dev/bin/readfish validate human_chr_selection.toml
2023-10-05 15:29:18,934 readfish command='validate'
2023-10-05 15:29:18,934 readfish log_file=None
2023-10-05 15:29:18,934 readfish log_format='%(asctime)s %(name)s %(message)s'
2023-10-05 15:29:18,934 readfish log_level='info'
2023-10-05 15:29:18,934 readfish no_check_plugins=False
2023-10-05 15:29:18,934 readfish no_describe=False
2023-10-05 15:29:18,934 readfish prom=False
2023-10-05 15:29:18,934 readfish toml='human_chr_selection.toml'
2023-10-05 15:29:18,934 readfish.validate eJydVk1v2zgQvetXEMqlxdryxyZAGyAHt0WKAk1TNNlTkBVoiZKIUKQiUonTX79vSEmW2zRo1/BBIkdvZt68GfKIXXV1zdunU3Z9efGZZUYXsmSFVIIVpmWt4GruZC3YlluRcaWkLmdM6FZmFR7JKDpi7tGwrGpNbayphWWv8MLWS8Z1ztar16zAFnOVYFVXc81KoWHGpGacWaDAWStKaXQCrOtK2v6V8aZREnjOMLiGC661UJbxrDXWesTHylCsyjxmTCiVRAOE2PG6wRYeQ1ZdK/KQVKc1xf4oXYUQ2be3yXGyYttO3e0zd8I6GHm8jxSvzJjjbSkc3LeC2UZkspCAzGUrMqeeKB+KyJlaBZx+AXA1UBiLEYiTZYwXeOD2dLo6s4B3M6FzPLVgjswokj6RGQyrdj1bzlbL5eyvOCYMvxQTbZ8KqsCa0h1Dm3n3AugIOHhB0iBy61+tzAVxwvvEZgyUbw1ICQHYUA5hBWu4c6LVoBJ84eta7gjeG0/TRki+qklmHzwHnr8NXJrCW20FKoUdofLAo9g1ikuNMPBhbbCSC8elGmBzk3U1UuCOBDEHWuVcY08XC2WMFYpvkxJ17LaJNAvINS+krRYUTFK5WpH7d710RTsqwaNFN2E1tcJRrW1Sdk3zdOurMv39639szuJgEY8UW2Y6oFaIRI8tIqglPpKhX5r3bXPoPChE81pEfdOdsTjXPG1XS5JjKt4k6/R4udw2Nj25q76nBbeONLHJ81ZA/T2j5eioz3FONQOLBe+UY7y3JiUFUyhENhkIXLi6WYSMFif4JdFgjFCeNyH/54jjHnm7pnMeVupcPsi844rmBWQTGhD/y6/Xny6/bD4jJu7bFVKivAcdZTQG7jvpBFMkQRLcN1GbB7xDE9T3ubR8SzrKxbYrU2U8UUo+iDQ4K+7joDFZamT//rDCNUbItML0/nKFvcW0wn6BlO0f5q3tc8FI8i4H56RSEFCgp3TmY++sSNhVZTqVU9MI6BQRnm+urjd+AGh2cfEpKnQq810KvSOxRQVKFjw3Wp4sPvTat4t30khNcwRpZRY6L+yiKv9+k2qTcuXAAk/K74nFuHRhA8MgvUjqWlLJLhtiA/X5ujmfVo5oDGl4N39G/+S7hhfk5ktXb5GgF6YvziAPsSP9vwpM0qEwUPl6fNsbEMNGq6fXkU4HnDN2HPng/LFwUGMUzQrxZ1PhiIOMJyvtPBw0IVA/fUaaw2n0QRRSS+dtkFfc6a0y2V2Mady0JhMij30KsXWmgSIzgVaA0GI/3DgBK0w8G0UksyPWf3RKMxGD0IBl7zarOn1HhNNo5o2jwwrzVRS06donoge7Nb8DqjZeSDkUahHZjMlEDMg+E20eB4d9wKfsn/DglUuMDAaHgQ+BDVZ9SFbcd6RqxETZ0jds/AZneEni8jn0NXc/HuX+3An3huFmkdux8s7fH/obAz2tkujmpi/O7W1Ec5KEh/tDSidzHNVSp73DM7ZCHliQdVczPYqw3+5J5CNf4yHGcxHVfLfHOSYcvnseJzQ0nUsjEMpB0WO6FTgeQRryxbghBYUJsUUvpRMXPPNjabhInLGb+Mv7dLlav10vk1V8SwX58bbhbyN7JqNwY0qNnxeH1Yvp+433QeE6Uov0x0TrL0Jebj3lZOI9JFGNg0L+CvBlRC9eh3vZFHvWX60cUwKHhd8afA3RFwV5G9ppmMO/HT3pRDq/CqQAPuTxPPQL2NSq/lu6L/YeLJIIm9AtGez9JBGmLjqCvIxDYPJ7MQltxmaazhqChOdnA/8NyIGWKeJP4jvA/gXiz7IPWqD7mQ16ZnvIyF/n0oNenFDyv3yEG+IeMvoPUvBL7w==
2023-10-05 15:29:18,937 readfish.validate Loaded TOML config without error
2023-10-05 15:29:18,937 readfish.validate Initialising Caller
2023-10-05 15:29:18,945 readfish.validate Caller initialised
2023-10-05 15:29:18,945 readfish.validate Initialising Aligner
2023-10-05 15:29:18,947 readfish.validate Aligner initialised
2023-10-05 15:29:18,948 readfish.validate Configuration description:
Region hum_test (control=False).
Region applies to section of flow cell (# = applied, . = not applied):
################################
################################
################################
################################
################################
################################
################################
################################
2023-10-05 15:29:18,948 readfish.validate Using the mappy plugin. Using reference: /home/adoni5/Documents/Bioinformatics/refs/hg38_no_alts.fa.gz.split/hg38_chr_M.mmi.
Region hum_test has targets on 1 contig, with 1 found in the provided reference.
This region has 2 total targets (+ve and -ve strands), covering approximately 100.00% of the genome.
If your toml file validates then run the following command:
readfish targets --toml <PATH_TO_TOML> --device <YOUR_DEVICE_ID> --log-file test.log --experiment-name human_select_testIn the terminal window you should see messages reporting the speed of mapping of the form:
2023-10-05 15:24:03,910 readfish.targets MinKNOW is reporting PHASE_MUX_SCAN, waiting for PHASE_SEQUENCING to begin. 2023-10-05 15:25:48,150 readfish._read_until_client Protocol phase changed to PHASE_SEQUENCING 2023-10-05 15:25:48,724 readfish.targets 0494R/0.5713s; Avg: 0494R/0.5713s; Seq:0; Unb:494; Pro:0; Slow batches (>1.00s): 0/1 2023-10-05 15:25:52,132 readfish.targets 0004R/0.1831s; Avg: 0249R/0.3772s; Seq:0; Unb:498; Pro:0; Slow batches (>1.00s): 0/2 2023-10-05 15:25:52,600 readfish.targets 0122R/0.2494s; Avg: 0206R/0.3346s; Seq:0; Unb:620; Pro:0; Slow batches (>1.00s): 0/3 2023-10-05 15:25:52,967 readfish.targets 0072R/0.2144s; Avg: 0173R/0.3046s; Seq:0; Unb:692; Pro:0; Slow batches (>1.00s): 0/4 2023-10-05 15:25:53,349 readfish.targets 0043R/0.1932s; Avg: 0147R/0.2823s; Seq:0; Unb:735; Pro:0; Slow batches (>1.00s): 0/5 2023-10-05 15:25:53,759 readfish.targets 0048R/0.2011s; Avg: 0130R/0.2688s; Seq:0; Unb:783; Pro:0; Slow batches (>1.00s): 0/6 2023-10-05 15:25:54,206 readfish.targets 0126R/0.2458s; Avg: 0129R/0.2655s; Seq:0; Unb:909; Pro:0; Slow batches (>1.00s): 0/7 2023-10-05 15:25:54,580 readfish.targets 0082R/0.2180s; Avg: 0123R/0.2595s; Seq:0; Unb:991; Pro:0; Slow batches (>1.00s): 0/8 2023-10-05 15:25:54,975 readfish.targets 0053R/0.2110s; Avg: 0116R/0.2542s; Seq:0; Unb:1,044; Pro:0; Slow batches (>1.00s): 0/9 2023-10-05 15:25:55,372 readfish.targets 0057R/0.2051s; Avg: 0110R/0.2492s; Seq:0; Unb:1,101; Pro:0; Slow batches (>1.00s): 0/10 2023-10-05 15:25:55,817 readfish.targets 0135R/0.2467s; Avg: 0112R/0.2490s; Seq:0; Unb:1,236; Pro:0; Slow batches (>1.00s): 0/11 2023-10-05 15:25:56,192 readfish.targets 0086R/0.2206s; Avg: 0110R/0.2466s; Seq:0; Unb:1,322; Pro:0; Slow batches (>1.00s): 0/12 2023-10-05 15:25:56,588 readfish.targets 0060R/0.2138s; Avg: 0106R/0.2441s; Seq:0; Unb:1,382; Pro:0; Slow batches (>1.00s): 0/13 2023-10-05 15:25:56,989 readfish.targets 0060R/0.2123s; Avg: 0103R/0.2418s; Seq:0; Unb:1,442; Pro:0; Slow batches (>1.00s): 0/14 2023-10-05 15:25:57,429 readfish.targets 0133R/0.2502s; Avg: 0105R/0.2424s; Seq:0; Unb:1,575; Pro:0; Slow batches (>1.00s): 0/15 2023-10-05 15:25:57,809 readfish.targets 0089R/0.2280s; Avg: 0104R/0.2415s; Seq:0; Unb:1,664; Pro:0; Slow batches (>1.00s): 0/16 2023-10-05 15:25:58,210 readfish.targets 0059R/0.2247s; Avg: 0101R/0.2405s; Seq:0; Unb:1,723; Pro:0; Slow batches (>1.00s): 0/17 ^C2023-10-05 15:25:58,238 readfish.targets Keyboard interrupt received, stopping readfish
WARNING |
|---|
Note: if these times are longer than the number of seconds specified in the break read chunk in the sequencing TOML, you will have performance issues. Contact us via github issues for support. |
This log is a little dense at first. Moving from left to right, we have:
[Date Time] [Logger Name] [Batch Stats]; [Average Batch Stats]; [Count commands sent]; [Slow Batch Info]
Using the provided log as an example:
On 2023-10-05 at 15:25:56,989, the Readfish targets command logged a batch of read signal:
- It saw 60 reads in the current batch.
- The batch took 0.2123 seconds.
- On average, batches are 103 reads, which are processed in 0.2418 seconds.
- Since the start, 0 reads were sequenced, 1,442 reads were unblocked, and 0 reads were asked to proceed.
- Out of 14 total batches processed, 0 were considered slow (took more than 1 second).
The important thing to note here is that the average batch time is less than the break read chunk time in the sequencing TOML. The slow batch section will show the number of batches that were slower than break reads. If the average is lower, or the slow batch count is high, you will have performance issues. Contact us via github issues for support.
If you are happy with the speed of mapping, move on to testing a selection.
Testing expected results from a selection experiment.
The only way to test readfish on a playback run is to look at changes in read length for rejected vs accepted reads. To do this:
Start a fresh simulation run using the bulkfile provided above.
Restart the readfish command (as above):
readfish targets --toml <PATH_TO_TOML> --device <YOUR_DEVICE_ID> --log-file test.log --experiment-name human_select_testAllow the run to proceed for at least 15 minutes (making sure you are writing out read data!).
After 15 minutes it should look something like this:
 If one zooms in on the unblock peak:
If one zooms in on the unblock peak:
 And if one zooms to exclude the unblock peak:
And if one zooms to exclude the unblock peak:
 NOTE: These simulations are also run on Apple Silicon - GPU platform performance may vary - please contact us via github issues for support.
NOTE: These simulations are also run on Apple Silicon - GPU platform performance may vary - please contact us via github issues for support.
Analysing results with readfish stats
Once a run is complete, it can be analysed with the readfish stats command.
HTML file output is optional.
readfish stats --toml <path/to/toml/file.toml> --fastq-directory <path/to/run/folder> --html <filename>
Readfish stats will use the initial experiment configuration to analyse the final sequence data and output a formatted table to the screen. The table is broken into two sections. For clarity these are shown individually below.
In the first table, the data is summarised by condition as defined in the TOML file. In this example we have a single Region - “hum_test”. The total number of reads is shown, along with the number of alignments broken down into On-Target and Off-Target. In addition, we show yield, median read length and a summary of the number of targets.
| Condition | Reads | Alignments | Yield | Median read lengths | Number of targets | Percent target | Estimated coverage | |||||||
| On-Target | Off-Target | Total | On-Target | Off-Target | Total | Ratio | On-target | Off-target | Combined | |||||
| hum_test | 112,058 | 819 (0.73%) | 111,239 (99.27%) | 112,058 | 9.27 Mb (5.49%) | 159.43 Mb (94.51%) | 168.69 Mb | 1:17.20 | 0 b | 896 b | 896 b | 2 | 3.60% | 0.08 X |
| On-Target | Off-Target | Total | On-Target | Off-Target | Total | Ratio | On-target | Off-target | Combined | |||||
| Condition | Reads | Alignments | Yield | Median read lengths | Number of targets | Percent target | Estimated coverage | |||||||
The lower portion of the table shows the data broken down by contig in the reference (and so can be very long if using a complex reference!). Again data are broken down by On and Off target. Read counts, yield, median and N50 read lengths are presented. Finally we estimate the proportion of reads on target and an estimate of coverage.
In this experiment, we were targeting chromosomes 20 and 21. As this is a playback run there is no effect on yield but you can see a clear effect on read length. The read length N50 and Median is higher for chromosomes 20 and 21 as expected. If running on more performant systems, the anticipated difference would be higher.
| Condition Name | hum_test | ||||||||||||||||||||
| Condition | Contig | Contig Length | Reads | Alignments | Yield | Median read lengths | N50 | Number of targets | Percent target | Estimated coverage | |||||||||||
| Mapped | Unmapped | Total | On-Target | Off-Target | Total | On-Target | Off-Target | Total | Ratio | On-target | Off-target | Combined | On-Target | Off-Target | Total | ||||||
| hum_test | chr1 | 248,956,422 | 10,015 | 0 | 10,015 | 6 (0.06%) | 10,009 (99.94%) | 10,015 | 48.65 Kb (0.37%) | 13.03 Mb (99.63%) | 13.08 Mb | 1:267.87 | 0 b | 891 b | 891 b | 0 b | 1.35 Kb | 1.35 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr2 | 242,193,529 | 8,825 | 0 | 8,825 | 9 (0.10%) | 8,816 (99.90%) | 8,825 | 47.36 Kb (0.36%) | 13.05 Mb (99.64%) | 13.09 Mb | 1:275.51 | 0 b | 894 b | 894 b | 0 b | 1.49 Kb | 1.49 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr3 | 198,295,559 | 8,005 | 0 | 8,005 | 6 (0.07%) | 7,999 (99.93%) | 8,005 | 193.03 Kb (1.73%) | 10.98 Mb (98.27%) | 11.17 Mb | 1:56.86 | 0 b | 893 b | 893 b | 0 b | 1.42 Kb | 1.42 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr4 | 190,214,555 | 7,381 | 0 | 7,381 | 30 (0.41%) | 7,351 (99.59%) | 7,381 | 861.07 Kb (7.29%) | 10.95 Mb (92.71%) | 11.81 Mb | 1:12.72 | 0 b | 917 b | 917 b | 0 b | 1.60 Kb | 1.60 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr5 | 181,538,259 | 7,545 | 0 | 7,545 | 5 (0.07%) | 7,540 (99.93%) | 7,545 | 50.70 Kb (0.50%) | 10.18 Mb (99.50%) | 10.23 Mb | 1:200.68 | 0 b | 896 b | 896 b | 0 b | 1.40 Kb | 1.40 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr6 | 170,805,979 | 5,808 | 0 | 5,808 | 9 (0.15%) | 5,799 (99.85%) | 5,808 | 116.44 Kb (1.35%) | 8.53 Mb (98.65%) | 8.65 Mb | 1:73.28 | 0 b | 905 b | 905 b | 0 b | 1.49 Kb | 1.49 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr7 | 159,345,973 | 6,383 | 0 | 6,383 | 2 (0.03%) | 6,381 (99.97%) | 6,383 | 26.06 Kb (0.29%) | 9.11 Mb (99.71%) | 9.14 Mb | 1:349.59 | 0 b | 895 b | 895 b | 0 b | 1.44 Kb | 1.44 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr8 | 145,138,636 | 5,208 | 0 | 5,208 | 1 (0.02%) | 5,207 (99.98%) | 5,208 | 285 b (0.00%) | 7.43 Mb (100.00%) | 7.43 Mb | 1:26061.60 | 0 b | 892 b | 892 b | 0 b | 1.44 Kb | 1.44 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr9 | 138,394,717 | 4,253 | 0 | 4,253 | 23 (0.54%) | 4,230 (99.46%) | 4,253 | 91.15 Kb (1.50%) | 6.00 Mb (98.50%) | 6.09 Mb | 1:65.85 | 0 b | 899 b | 899 b | 0 b | 1.46 Kb | 1.46 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr10 | 133,797,422 | 4,424 | 0 | 4,424 | 15 (0.34%) | 4,409 (99.66%) | 4,424 | 95.02 Kb (1.37%) | 6.86 Mb (98.63%) | 6.95 Mb | 1:72.16 | 0 b | 915 b | 915 b | 0 b | 1.56 Kb | 1.56 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr11 | 135,086,622 | 5,349 | 0 | 5,349 | 1 (0.02%) | 5,348 (99.98%) | 5,349 | 287 b (0.00%) | 6.89 Mb (100.00%) | 6.89 Mb | 1:23997.50 | 0 b | 896 b | 896 b | 0 b | 1.35 Kb | 1.35 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr12 | 133,275,309 | 5,508 | 0 | 5,508 | 3 (0.05%) | 5,505 (99.95%) | 5,508 | 2.63 Kb (0.03%) | 7.59 Mb (99.97%) | 7.59 Mb | 1:2888.96 | 0 b | 893 b | 893 b | 0 b | 1.40 Kb | 1.40 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr13 | 114,364,328 | 3,414 | 0 | 3,414 | 8 (0.23%) | 3,406 (99.77%) | 3,414 | 85.71 Kb (1.80%) | 4.69 Mb (98.20%) | 4.77 Mb | 1:54.67 | 0 b | 900 b | 900 b | 0 b | 1.43 Kb | 1.43 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr14 | 107,043,718 | 3,541 | 0 | 3,541 | 12 (0.34%) | 3,529 (99.66%) | 3,541 | 244.18 Kb (4.79%) | 4.86 Mb (95.21%) | 5.10 Mb | 1:19.90 | 0 b | 892 b | 892 b | 0 b | 1.42 Kb | 1.42 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr15 | 101,991,189 | 3,033 | 0 | 3,033 | 3 (0.10%) | 3,030 (99.90%) | 3,033 | 4.29 Kb (0.11%) | 3.79 Mb (99.89%) | 3.80 Mb | 1:883.07 | 0 b | 867 b | 867 b | 0 b | 1.31 Kb | 1.31 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr16 | 90,338,345 | 3,276 | 0 | 3,276 | 1 (0.03%) | 3,275 (99.97%) | 3,276 | 1.97 Kb (0.04%) | 4.51 Mb (99.96%) | 4.51 Mb | 1:2294.28 | 0 b | 900 b | 900 b | 0 b | 1.41 Kb | 1.41 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr17 | 83,257,441 | 3,378 | 0 | 3,378 | 10 (0.30%) | 3,368 (99.70%) | 3,378 | 16.81 Kb (0.36%) | 4.72 Mb (99.64%) | 4.73 Mb | 1:280.52 | 0 b | 907 b | 907 b | 0 b | 1.43 Kb | 1.43 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr18 | 80,373,285 | 3,158 | 0 | 3,158 | 3 (0.09%) | 3,155 (99.91%) | 3,158 | 186.59 Kb (4.06%) | 4.41 Mb (95.94%) | 4.59 Mb | 1:23.61 | 0 b | 899 b | 899 b | 0 b | 1.47 Kb | 1.47 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr19 | 58,617,616 | 2,110 | 0 | 2,110 | 0 (0.00%) | 2,110 (100.00%) | 2,110 | 0 b (0.00%) | 2.53 Mb (100.00%) | 2.53 Mb | 0:0.00 | 0 b | 857 b | 857 b | 0 b | 1.27 Kb | 1.27 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chr20 | 64,444,167 | 370 | 0 | 370 | 370 (100.00%) | 0 (0.00%) | 370 | 3.60 Mb (100.00%) | 0 b (0.00%) | 3.60 Mb | 1:0.00 | 0 b | 2.88 Kb | 2.88 Kb | 0 b | 32.28 Kb | 32.28 Kb | 1 | 100.00% | 0.06 X |
| hum_test | chr21 | 46,709,983 | 265 | 0 | 265 | 265 (100.00%) | 0 (0.00%) | 265 | 3.06 Mb (100.00%) | 0 b (0.00%) | 3.06 Mb | 1:0.00 | 0 b | 2.63 Kb | 2.63 Kb | 0 b | 33.54 Kb | 33.54 Kb | 1 | 100.00% | 0.07 X |
| hum_test | chr22 | 50,818,468 | 1,741 | 0 | 1,741 | 28 (1.61%) | 1,713 (98.39%) | 1,741 | 421.99 Kb (14.61%) | 2.47 Mb (85.39%) | 2.89 Mb | 1:5.85 | 0 b | 922 b | 922 b | 0 b | 1.63 Kb | 1.63 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chrM | 16,569 | 19 | 0 | 19 | 0 (0.00%) | 19 (100.00%) | 19 | 0 b (0.00%) | 16.82 Kb (100.00%) | 16.82 Kb | 0:0.00 | 0 b | 774 b | 774 b | 0 b | 1.11 Kb | 1.11 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chrX | 156,040,895 | 5,636 | 0 | 5,636 | 5 (0.09%) | 5,631 (99.91%) | 5,636 | 3.19 Kb (0.04%) | 7.46 Mb (99.96%) | 7.46 Mb | 1:2336.71 | 0 b | 905 b | 905 b | 0 b | 1.38 Kb | 1.38 Kb | 0 | 0.00% | 0.00 X |
| hum_test | chrY | 57,227,415 | 116 | 0 | 116 | 4 (3.45%) | 112 (96.55%) | 116 | 117.28 Kb (27.65%) | 306.90 Kb (72.35%) | 424.19 Kb | 1:2.62 | 0 b | 989 b | 989 b | 0 b | 28.59 Kb | 28.59 Kb | 0 | 0.00% | 0.00 X |
| hum_test | unmapped | 0 | 0 | 3,297 | 3,297 | 0 (0.00%) | 3,297 (100.00%) | 3,297 | 0 b (0.00%) | 9.10 Mb (100.00%) | 9.10 Mb | 0:0.00 | 0 b | 508 b | 508 b | 0 b | 16.81 Kb | 16.81 Kb | 0 | 0.00% | 0.00 X |