Developer’s guide¶
If you want to contribute some code to readfish, please check out the contributors guide on GitHub.
Pre-commit¶
In order to enforce coding style, we use pre-commit.
Installation¶
pip install pre-commit
cd readfish
pre-commit install
pre-commit run --all-files
In order to run the checks automatically when committing/checking out branches, run the following command
pre-commit install -t pre-commit -t post-checkout -t post-merge
Readfish Installation¶
We use pyproject.toml to handle building, packaging and installation, along with hatch.
Installation candidates are listed in the pyproject.toml like so
[project.optional-dependencies]
# Development dependencies
docs = ["sphinx-copybutton", "furo", "myst-parser", "faqtory"]
tests = ["pytest"]
dev = ["readfish[all,docs,tests]"]
# Running dependencies, this is a little bit clunky but works for now
mappy = ["mappy"]
mappy-rs = ["mappy-rs"]
dorado = ["ont-pybasecall-client-lib"]
all = ["readfish[mappy,mappy-rs,dorado]"]
An example install command would be pip install readfish[all].
An example conda development environment is specified as:
name: readfish_dev
channels:
- bioconda
- conda-forge
- defaults
dependencies:
- python=3.10
- pip
- pip:
- -e .[dev]
This file is included here: development.yml, and can be installed in the main readfish repository like so:
With mamba:
mamba env create -f docs/development.yml
With conda:
conda env create -f docs/development.yml
Readfish versioning¶
Readfish uses calver for versioning. Specifically the format should be
YYYY.MINOR.MICRO.Modifier, where MINOR is the feature addition, MICRO is any hotfix/bugfix, and Modifier is the modifier (e.g. rc for release candidate, dev for development, empty for stable).
Changelog¶
We are generally trying to follow the guidance here. https://keepachangelog.com/en/1.0.0/
Notably we should correctly update the Unreleased section for things added in the PRs that are inbetween releases.
If possible, link the PR that introduced the change, and add a brief description of the change.
Adding a simulated position for testing¶
The following steps should all happen with a configuration (test) flow cell inserted into the target device. A simulated device can also be created within MinKNOW, following these instructions. This assumes that you are runnning MinKNOW locally, using default ports. If this is not the case a developer API token is required on the commands as well, as well as setting the correct port.
If no test flow cell is available, a simulated device can be created within MinKNOW, following the below instructions.
Adding a simulated position for testing
Linux
In the readfish virtual environment we created earlier:
See help
python -m minknow_api.examples.manage_simulated_devices --helpAdd Minion position
python -m minknow_api.examples.manage_simulated_devices --add MS00000Add PromethION position
python -m minknow_api.examples.manage_simulated_devices --prom --add S0Mac
In the readfish virtual environment we created earlier:
See help
python -m minknow_api.examples.manage_simulated_devices --helpAdd Minion position
python -m minknow_api.examples.manage_simulated_devices --add MS00000Add PromethION position
python -m minknow_api.examples.manage_simulated_devices --prom --add S0
As a back up it is possible to restart MinKNOW with a simulated device. This is done as follows:
Stop
minknowOn Linux:
cd /opt/ont/minknow/bin sudo systemctl stop minknow
Start MinKNOW with a simulated device
On Linux
sudo ./mk_manager_svc -c /opt/ont/minknow/conf --simulated-minion-devices=1 &
You may need to add the host 127.0.0.1 in the MinKNOW UI.
Setting up a test playback experiment¶
Previously to set up Playback using a pre-recorded bulk FAST5 file, it was necessary to edit the sequencing configuration file that MinKNOW uses. This is currently no longer the case. The “old method” steps are left after this section for reference only or if the direct playback from a bulk file option is removed in future.
To start sequencing using playback, simply begin setting up the run in the MinKNOW UI as you would usually. Under Run Options you can select Simulated Playback and browse to the downloaded Bulk Fast5 file.
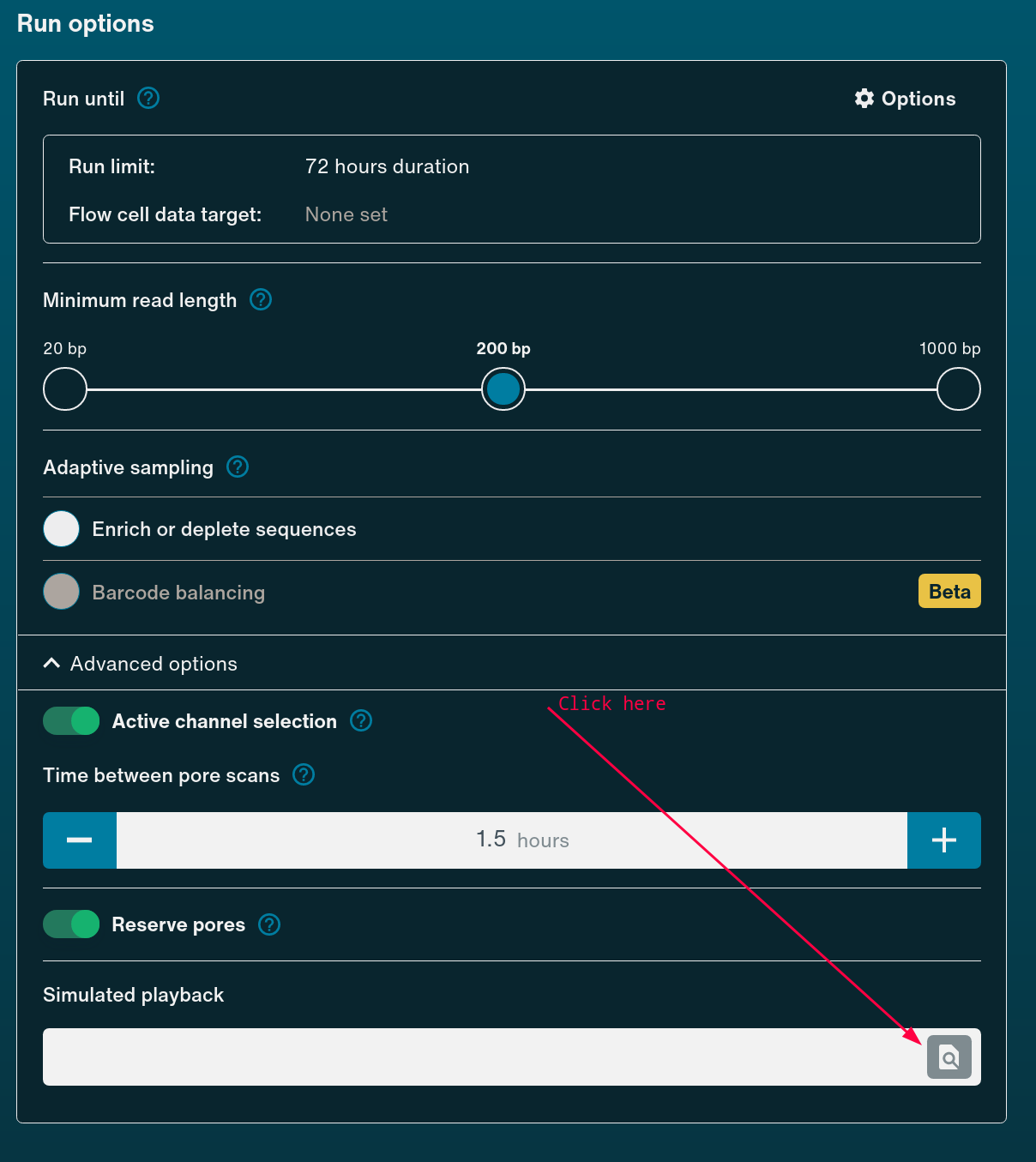
Viewing Documentation¶
Aside from a traditional README.md, readfish uses sphinx to create documentation.
All documentation is kept in the readfish/docs directory.
A live version is available at https://looselab.github.io/readfish/.
With an activated development readfish environment (includes docs dependencies), run the following to view the documentation:
cd docs
make html
cd _build/html
python -m http.server 8080
The documentation HTML should now be available at http://0.0.0.0:8080/.
Adding to Documentation¶
The main file for configuring sphinx is conf.py.
This file should not need altering, it sets things up for activated sphinx extensions, auto generated API documentation, building etc.
sphinx builds from markdown files (amongst others), with index.md as the master_doc page that other pages are included into.
If another markdown file is added, it should be included into the {toc_tree} directive in index.md.
Otherwise documentation should be written using standard markdown, excepting API documentation, which is written in ReStructured Text.
An example of this is readfish.console.rst, which auto generates a documentation page for all entry points in readfish.
However, the entry points to be documented must be added by a deVeLoPEr.
Entry points¶
Entry points are the subcommands that readfish runs.
These are found in src/readfish/entry_points.
Excepting validate.py, ideally these wrap targets.py, which performs the bare minimum read_until loop, calling the Aligner and Caller plugins.
If adding an entry point, they must also be included in src/readfish/_cli_base.py,adding to this cmds list
cmds = [
("targets", "targets"),
("barcode-targets", "targets"),
("unblock-all", "unblock_all"),
("validate", "validate"),
("stats", "stats"),
]
# Entry point is imported during runtime, and added as a sub command to readfish
for cmd, module in cmds:
_module = importlib.import_module(f"readfish.entry_points.{module}")
_parser = subparsers.add_parser(cmd, help=_module._help)
for *flags, opts in _module._cli:
_parser.add_argument(*flags, **opts)
_parser.set_defaults(func=_module.run)
Plugin modules¶
Plugin modules are designed to make readfish more modular.
Each Plugin module should inherit an Abstract Base Class (ABC), which define methods which MUST be present on the plugin.
Currently, two types of plugins can be written
Callerplugins (must inheritCallerABC), which wrap a base caller for calling chunksAlignerplugins (must inheritAlignerABC), which wrap an aligner for making decisions on base called chunks.
These ABCs are defined in abc.py.
This means that the methods which are used in targets.py are present, so plugins can be swapped out at run time, and function as standardised self-contained interfaces to different external tools.
If a plugin module is written and not included in readfish package, but another, it is possible to include it by passing the path in the toml.
For example:
Plugins are loaded as instances of _PluginModules - see source of _PluginModules for more details.
- class readfish._config._PluginModule(name, parameters)[source]
A plugin module
[caller_settings.bar.foo]
This would try to load the foo module from the bar package.
[caller_settings.readfish.plugins.dorado]
This would load the readfish dorado Caller plugin explicitly.
There are instances of “builtin” plugins, which are the included mappy, mappy_rs and dorado plugins.
See the source of readfish._config._PluginModule.load_module for more details.
- _PluginModule.load_module(override=False)[source]
Load a plugin module with the given name.
If the module is a built-in plugin (as specified in the builtins dictionary), it is loaded from the readfish.plugins package. Otherwise, it is loaded using the importlib library.
- Parameters:
override – If True, the built-in module names are ignored. Default is False.
- Returns:
The loaded module.
- Raises:
ModuleNotFoundError – If the plugin module cannot be found or loaded.
Note that this method is intended to be used as part of a plugin system, where plugin modules are loaded dynamically at runtime. The builtins dictionary maps the names of built-in plugins to the actual module names, and is used to avoid having to specify the full module name when loading a built-in plugin. If override=True, the builtin module names are ignored.
for a list of built in modules. This is how these plugins can be loaded, without passing an absolute import path to them.
Validation is left to the author of any plugins which inherits from the Aligner ABC. Things we suggest that are validated:
required keys - keys that must be present in the TOML
correctly typed values - Values that have been passed in ar eof the correct instance
available input files - Check the existence of paths
writable outputs - Check permissions on output files
Let’s dissect the structure of _mappy.py. In this case, _mappy.py defines shared behaviour between two Plugins, mappy and mappy_rs.
These files are in essence identical, however they use a different Aligner( mappy and mappy-rs respectively ),
and so are in separate files to allow the User to have a discrete choice between them when loading the plugin via the TOML file.
Here we define an Aligner class.
It is Required that the class name be Aligner for Aligner plugins and Caller for Caller plugins.
We inherit the AlignerABC class, which provides the required methods for an Aligner.
class _Aligner(AlignerABC):
"""
A wrapper class for mappy.Aligner providing an interface to either `mappy` or `mappy-rs` aligner implementations.
:param Aligners mappy_impl: Specifies which mappy implementation to use, represented by an `Aligners` enum instance.
:param Optional[str] debug_log: Specifies the file to log debug information to. If `None`, no logging is performed.
:param kwargs: Additional keyword arguments to be passed to the mappy aligner constructor.
:raises ValueError: When an invalid `mappy_impl` value is provided.
Example:
.. code-block:: python
aligner = _Aligner(mappy_impl=Aligners.C_MAPPY, debug_log="debug.log", preset="map-ont")
.. note::
The actual aligner instance is created during initialization based on the provided `mappy_impl`, and is accessible via the `aligner` attribute of the instance.
init()¶
Firstly, as with most python classes, we initialise the class.
We do not need to call super().init() as we aren’t actually using the initialised AlignerABC just the inherited methods.
"""
def __init__(self, mappy_impl: Aligners, debug_log: Optional[str] = None, **kwargs):
# Variant of the mappy aligner to use - `mappy` or `mappy_rs`
self.mappy_impl = mappy_impl
# Setup logger with the provided debug_log file, or a null logger if no file is provided.
self.logger = setup_logger(__name__, log_file=debug_log)
self.aligner_params = kwargs
# Validate the provided arguments will create a valid Aligner.
self.validate()
# Import selected aligner and Initialise it
if self.mappy_impl is Aligners.C_MAPPY:
import mappy
self.aligner = mappy.Aligner(**self.aligner_params)
elif self.mappy_impl is Aligners.MAPPY_RS:
import mappy_rs
self.aligner = mappy_rs.Aligner(**self.aligner_params)
threads = self.aligner_params.get("n_threads", 1)
self.aligner.enable_threading(threads)
else:
validate()¶
This next method is important. validate()
The validate function is intended to be called in the init method, before the actual Aligner or Caller is initialised. The contents of this method are left up to the author, however we suggest that people check for the things listed above.
The purpose of validate is to check that the given parameters will create a valid Aligner or Caller. For example, in the dorado.py Caller plugin, we check the permissions of the provided dorado socket. If these are insufficient, dorado only errors out after a 10 minute timeout. However if this is caught in validate, everyone ends up being left a lot happier.
def validate(self) -> None:
"""
Check that this aligner can be initialised without any issues. Catches any problems and raises helpful errors.
Currently checks:
1. that the Reference (fn_idx_in) exists, IF one is provided
2. That the reference is an .mmi file, or a FASTA or FASTQ, either uncompressed or Gzipped, IF a fn_idx_in is provided.
:return: None, if the Aligner is setup with valid paths and permissions
"""
index: str = self.aligner_params["fn_idx_in"]
file_extensions = [".fasta", ".fna", ".fsa", ".fa", ".fastq", ".fq"]
file_extensions.extend([f"{f}.gz" for f in file_extensions])
file_extensions.append(".mmi")
if all((not Path(index).is_file(), index)):
raise FileNotFoundError(f"{index} does not exist")
if not any(index.lower().endswith(suffix) for suffix in file_extensions):
raise RuntimeError(
f"Provided index file appears to be of an incorrect type - should be one of {file_extensions}"
)
def disconnect(self) -> None:
disconnect()¶
Required disconnect method - we don’t have any clean up to do so we just return.
@property
initialised()¶
Required initialised`` method. In this case, we are initialised if the Aligner has an index loaded - which is when a mappy.Alignerormappy_rs.Alignerevaluates totrue`
"""Is the mappy Aligner initialised?
If ``False`` the ``Aligner`` is unlikely to work for mapping.
"""
return bool(self.aligner)
def map_reads(self, basecall_results: Iterable[Result]) -> Iterable[Result]:
map_reads()¶
Required function - map_reads.
This is required, and takes in an iterable of the Results from the Caller plugin.
The basecalled data present on the on the result is aligned through one of the two aligner wrapper functions
below, which set the alignments returned on the Result for this chunk.
The intention here is to allow the Caller plugin to be agnostic of the aligner used, and the Aligner plugin to be agnostic of the caller used,
as long as the Result object is properly populated.
Maps an iterable of base-called data using either the C `mappy` or `mappy-rs` Rust implementation,
based on the `mappy_impl` provided during the instantiation of the class.
:param Iterable[Result] basecall_results: An iterable of basecalled read results to be mapped.
:return: An iterable of mapped read results.
:raises NotImplementedError: If the aligner is not configured (i.e., if `mappy_impl` is neither `Aligners.MAPPY_RS` nor `Aligners.C_MAPPY`).
Example:
.. code-block:: python
mapped_results = aligner.map_reads(basecall_results)
.. note::
This method logs the mapping results or `UNMAPPED_PAF` for unmapped reads,
and the actual mapping is performed by `_c_mappy_wrapper` or `_rust_mappy_wrapper` methods based on the `mappy_impl`.
"""
# Map with MAPPY_RS
if self.mappy_impl is Aligners.MAPPY_RS:
iter_ = self._rust_mappy_wrapper(basecall_results)
# Map with MAPPY
elif self.mappy_impl is Aligners.C_MAPPY:
iter_ = self._c_mappy_wrapper(basecall_results)
else:
raise NotImplementedError("Aligner not configured")
# Iterate over the results and log them, then yield, now with Alignment data
# If there were alignments
for result in iter_:
paf_info = f"{result.read_id}\t{len(result.seq)}"
for al in result.alignment_data:
self.logger.debug(f"{paf_info}\t{al}")
if not result.alignment_data:
self.logger.debug(f"{paf_info}\t{UNMAPPED_PAF}")
yield result
def _c_mappy_wrapper(self, basecalls: Iterable[Result]) -> Result:
describe()¶
Our final required function describe. The contents of the description string is left to the developer of any plugins - all that need be returned is a string
describing what the aligner is seeing before it starts. This will be logged in the readfish log file and MinKNOW logs,
so things to bear in mind are things that will be useful when retrospectively analysing the run.
This function is called in targets.py to be logged to the terminal.
This is a more complicated function, so I’ve commented details more thoroughly below.
"""
Provides a human-readable description of the mappy Aligner plugin instance. This description
includes details about the plugin, the reference used, target regions, and barcodes, making it useful
for logging and debugging purposes.
:param list[Region] regions: A list of regions to be described.
:param list[Barcode] barcodes: A list of barcodes to be described.
:return: A formatted string containing detailed, human-readable information about the regions, barcodes,
and the state of the Aligner instance.
:raises SystemExit: If there are contigs listed as targets but not found in the target reference.
Example:
.. code-block:: python
description = aligner.describe(regions, barcodes)
print(description)
.. note::
The description generated by this method is intended for logging to readfish and MinKNOW and provides
insights into the configuration and status of the aligner instance, reference used, and target specifics.
If the Aligner is not initialised yet, it will return a message indicating that no reference has been
provided, and the Aligner needs initialisation before proceeding.
"""
# List to store strings, that will be concatenated at the end and returned.
description = []
# Check we have a reference.
if self.initialised:
description.append(
f"Using the {self.mappy_impl.value} plugin. Using reference: {(self.aligner_params['fn_idx_in'])}.\n"
)
# All the contig names that are in the reference.
seq_names = set(self.aligner.seq_names)
# Get total seq length of the reference.
genome = get_contig_lengths(self.aligner)
# Ref len for tow strands
ref_len = sum(genome.values()) * 2
# Print out for each region and barcode. Zip an identifying string in with the class containing the Condition data.
for condition, region_or_barcode_str in chain(
zip(regions, repeat("Region", len(regions))),
zip(barcodes.values(), repeat("Barcode", len(barcodes))),
):
# Create a set of all contigs listed as targets so contigs are unique
unique_contigs = set(condition.targets._targets[Strand.forward].keys())
unique_contigs.update(
set(condition.targets._targets[Strand.reverse].keys())
)
num_unique_contigs = len(unique_contigs)
# More than one contig should be plural!
pluralise = {1: ""}.get(num_unique_contigs, "s")
num_in_ref_contigs = len(unique_contigs & seq_names)
# Check all targets are in the reference.
num_not_in_ref_contigs = len(unique_contigs - seq_names)
# NOTE - we raise an error if we have a contig in the targets that is not in the reference.
warn_not_found = (
f"NOTE - The following {num_not_in_ref_contigs} {'contigs are listed as targets but have' if num_not_in_ref_contigs > 1 else 'contig is listed as a target but has'} not been found on the target reference:\n {nice_join(sorted(unique_contigs - seq_names), conjunction='and')}"
if num_not_in_ref_contigs
else ""
)
if warn_not_found:
raise SystemExit(warn_not_found)
# Calculate some fun stats - this could just count from the `iter_targets` method, but this is more fun.
num_targets = count_dict_elements(condition.targets._targets)
ref_covered = _summary_percent_reference_covered(
ref_len, condition.targets, genome
)
# Append a nicely formatted summation for each region or barcode
description.append(
f"""{region_or_barcode_str} {condition.name} has targets on {num_unique_contigs} contig{pluralise}, with {num_in_ref_contigs} found in the provided reference.
This {region_or_barcode_str.lower()} has {num_targets} total targets (+ve and -ve strands), covering approximately {ref_covered:.2%} of the genome.\n"""
)
# Join the list of strings together with newlines and return.
return "\n".join(description)
return "Aligner is not initialised yet. No reference has been provided, readfish will not proceed until the Aligner is initialised."
_c_mappy_wrapper()¶
Just a look at the _c_mappy_wrapper - a simple None required function that maps using mappy and returns mappy.Alignments
A private method to map an iterable of base-called data using the C minimap2 `mappy` implementation.
:param Iterable[Result] basecalls: An iterable of basecalled read results to be mapped.
:return: A generator yielding mapped read results.
"""
for result in basecalls:
result.alignment_data = list(self.aligner.map(result.seq))
yield result
def _rust_mappy_wrapper(self, basecalls: Iterable[Result]) -> Result: