ReadFish Multiplatform Update
Alex Payne 02 Dec 2020
Up until now ReadFish, and selective sequencing, have been limited to users with both a linux OS and an NVIDIA GPU. In reality this requires a fairly large box - be it a GridION or a desktop able to support an NVIDIA 1080 GPU or above. However, it would be ideal to be able to run selective sequencing on a laptop - even if it needs to be a moderately powerful device. This was already possible with ReadFish if you have a beefy NVIDIA GPU but many notable laptops cough Apple cough don’t support this at this time. Quite quickly after releasing our preprint people suggested using a CPU base caller:
Or just do CPU basecalling ;) https://t.co/ICgitgG4Hj
— Vlado Boza (@bozavlado) February 6, 2020
Given the way we developed ReadFish, it is relatively straightforward to use different components. We therefore decided to implement an alternative base calling strategy using DeepNano-Blitz instead of Guppy. As a consequence we have been able to implement selective sequencing on computers without GPU and running Linux, macOS and even Windows. This brings the power of selective sequencing to laptops (with a few caveats below).
This also enabled @mattloose to spend a happy weekend running selective sequencing on a human genome at home.
Results
So far we’ve run using DeepNano-Blitz on macOS, Windows (WSL), GridION, and Linux. In all cases we have seen enrichment comparable to running ReadFish using GPU accelerated base calling. A key caveat is that our DeepNano-Blitz implementation is currently non-adaptive; that is, it can enrich or deplete specific genomes or targets but this implementation does not automatically monitor reads as they are written or change targets during the experiment.
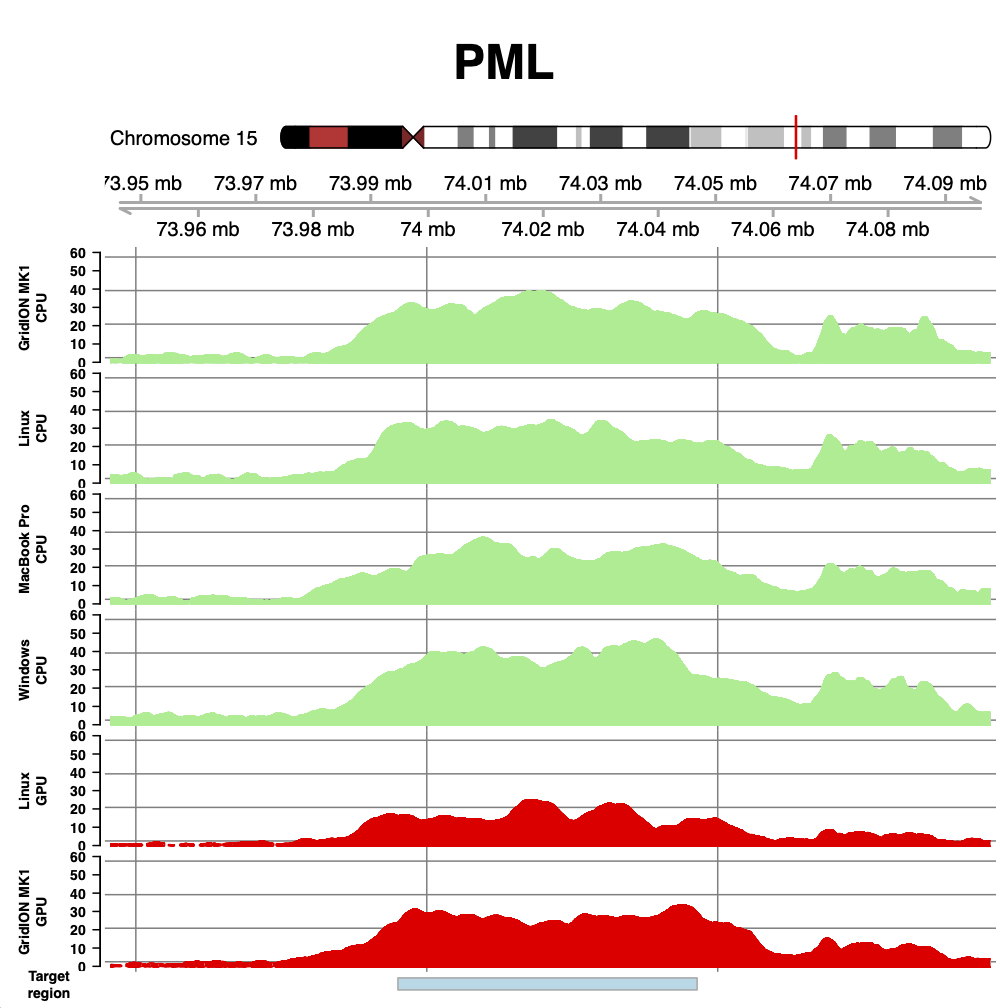
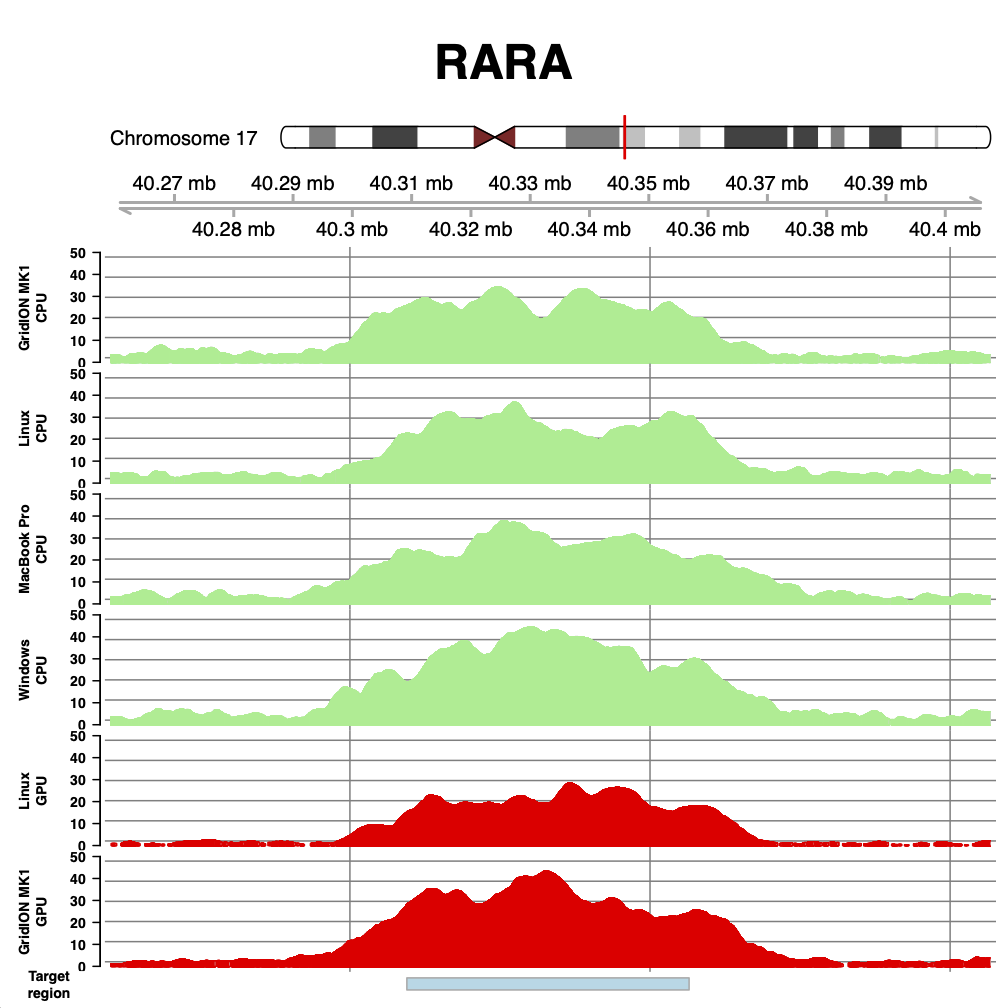
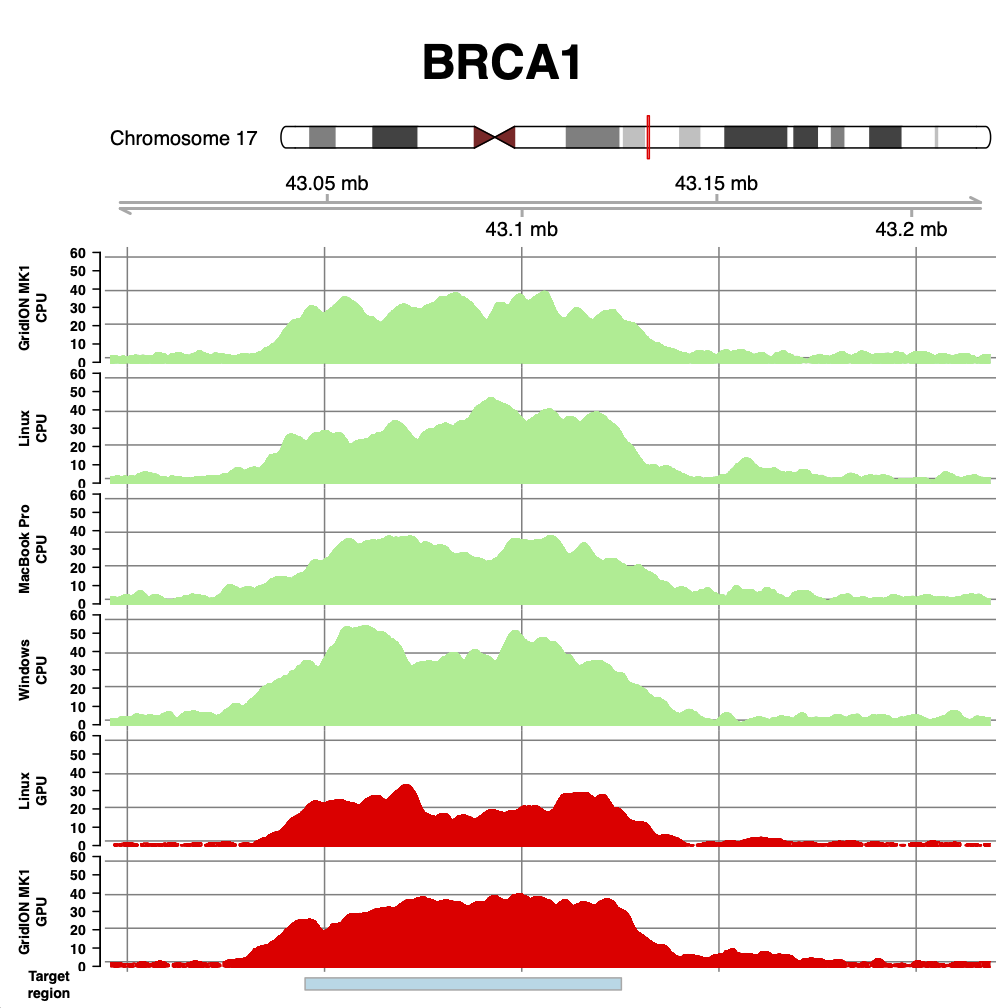
With that cleared up - what do we see? The figures below show coverage over BRCA1, PML and RARA, for runs on the GridION Mk1 CPU, a linux box we found in the lab, a MacBook Pro (2018 - 3.1 GHz i7), and a windows desktop running Windows Subsystem for Linux (windows 10). We also show two runs from our paper for comparison - the GridION Mk1 and the same linux box using an NVIDIA GTX 1080 GPU.
As can be seen, the coverage obtained with DeepNano-Blitz ReadFish is comparable (and on windows better!) than that seen on our original experiments.
In the plot below you can choose a chromosome and look at the distribution of coverage for each target. Broadly speaking this is pretty uniform although a few targets have pseudogenes that could be cleaned up in future (for example see chromosome 4).
The difference in coverage is largely down to down to yield for each flow cell. But there are some subtle differences which become apparent. Compare the GPU and CPU coverage on the GridION. The GPU experiment had much lower yield, but the median read length was shorter. Similarly, the Linux GPU has very low yield, but also one of the best median read lengths we have seen. The take home here is that speed is everything for efficient read until experiments. The faster you can unblock reads you don’t wish to see, the better (and flushing your flow cell can help recover capacity). Thus the windows run shown here, at 17 Gb, uses brute force of yield to compensate for the longer reads observed. There is still room for tuning the windows based setup.
| GridION MK1 GPU | GridION MK1 CPU | Linux GPU | Linux CPU | MacBook Pro CPU | Windows CPU | |
|---|---|---|---|---|---|---|
| Mean Read Length | 735.6 | 878.9 | 683.7 | 771.2 | 1085.0 | 1146.9 |
| Median Read Length | 423.0 | 662.0 | 402.0 | 564.0 | 745.0 | 823.0 |
| Yield | 9.08 Gb | 14.93 Gb | 5.90 Gb | 14.31 Gb | 14.03 Gb | 17.27 Gb |
| Mean Coverage | 31.30 | 29.78 | 19.11 | 27.78 | 29.08 | 34.47 |
| Coverage Std | 5.54 | 5.30 | 3.23 | 5.09 | 5.24 | 6.62 |
| Flushes | 2 | 2 | 2 | 2 | 2 | 3 |
Hardware Requirements
To run everything from one laptop (or desktop) we recommend at least 16 Gb of RAM, 8 processing cores and an SSD. The processor needs to be one of intel i7 or Xeon.
ReadFish will use four cores for base calling and mapping an entire flow cell in real time, leaving the remainder of the CPU power for MinKNOW. We have not run on an infinite number of systems and your mileage may vary, but DeepNano-Blitz provides a number of different models that you can try.
Caveats
This branch of ReadFish is under development and provided as is for you to try out.
Currently this is only compatible with R9.4 data on DNA. This contrasts with Guppy ReadFish, which can handle RNA as well as DNA and any currently shipping flow cell type from ONT.
We have not yet implemented real-time base calling for the downstream reads - so any of our tools that look at base called reads to inform the choice of reads for sequencing will not currently work.
So how do I set this up?
To get started you will need to download the pre-built DeepNano-Blitz installers:
If you run into issues accessing or installing these, please contact us.
On Windows we use Windows Subsytem Linux (WSL) to run ReadFish. To setup WSL see the Microsoft documentation. Once this is installed you may need to install or upgrade the Python3 version to be one of 3.6, 3.7, or 3.8; now continue the install using the instructions for Linux.
Setup a virtual environment:
python3 -m venv venv
source ./venv/bin/activate
pip install --upgrade pip wheel
Install DeepNano-Blitz:
Choose the .whl file corresponding to your OS and python version. E.g:
pip install deepnano2-0.1-cp36-cp36m-linux_x86_64.whl
Install ReadFish:
pip install git+https://github.com/nanoporetech/read_until_api@v3.0.0
pip install git+https://github.com/LooseLab/readfish@caller_refactor
# Install ont_pyguppy_client_lib that matches
# your guppy server version. E.G.
pip install ont_pyguppy_client_lib==4.0.11
Setting the CPU caller
To use DeepNano-Blitz for base calling the caller_settings table in the
experiment TOML file must contain the network_type parameter. For more
information on the available parameters see TOML.md.